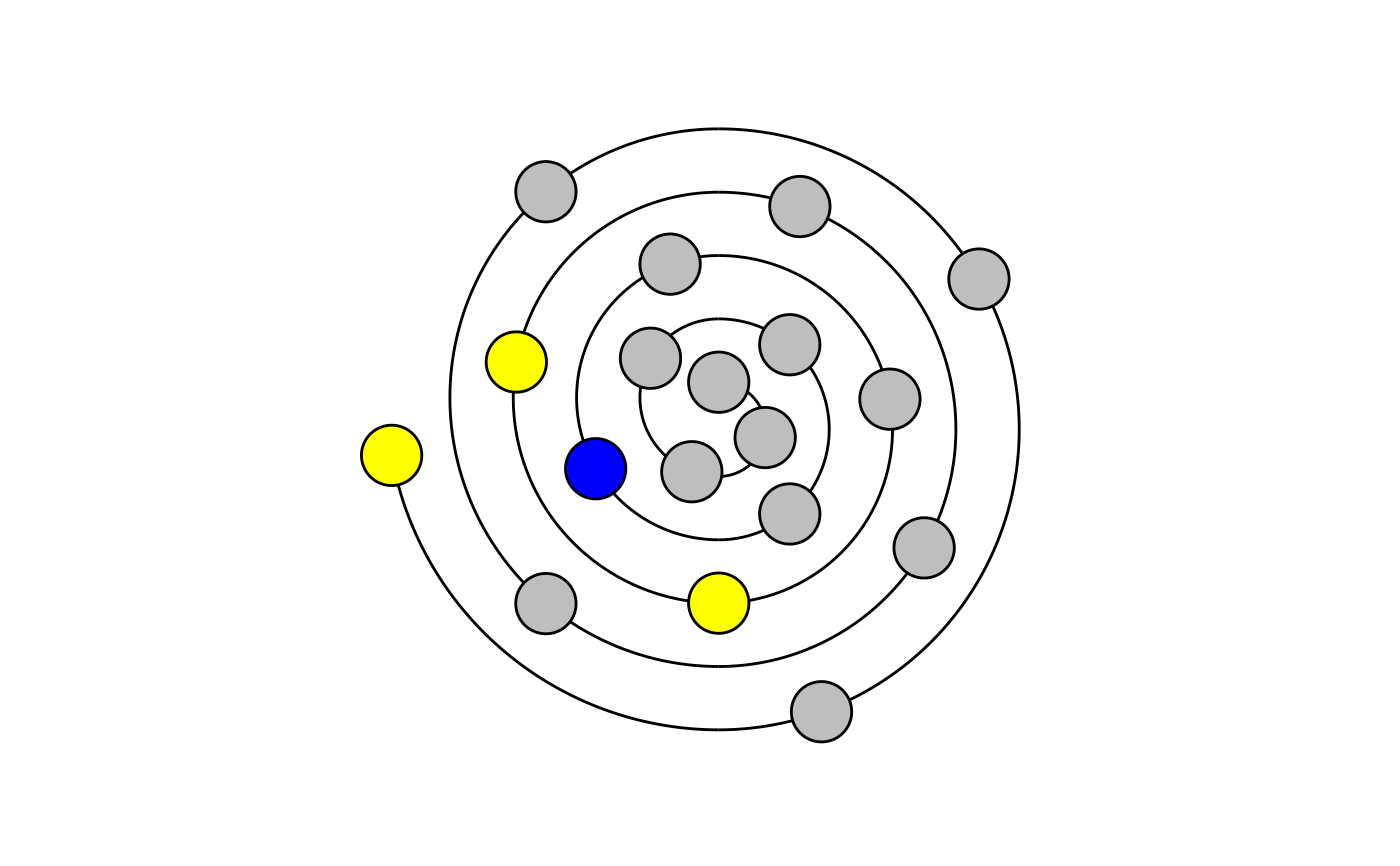
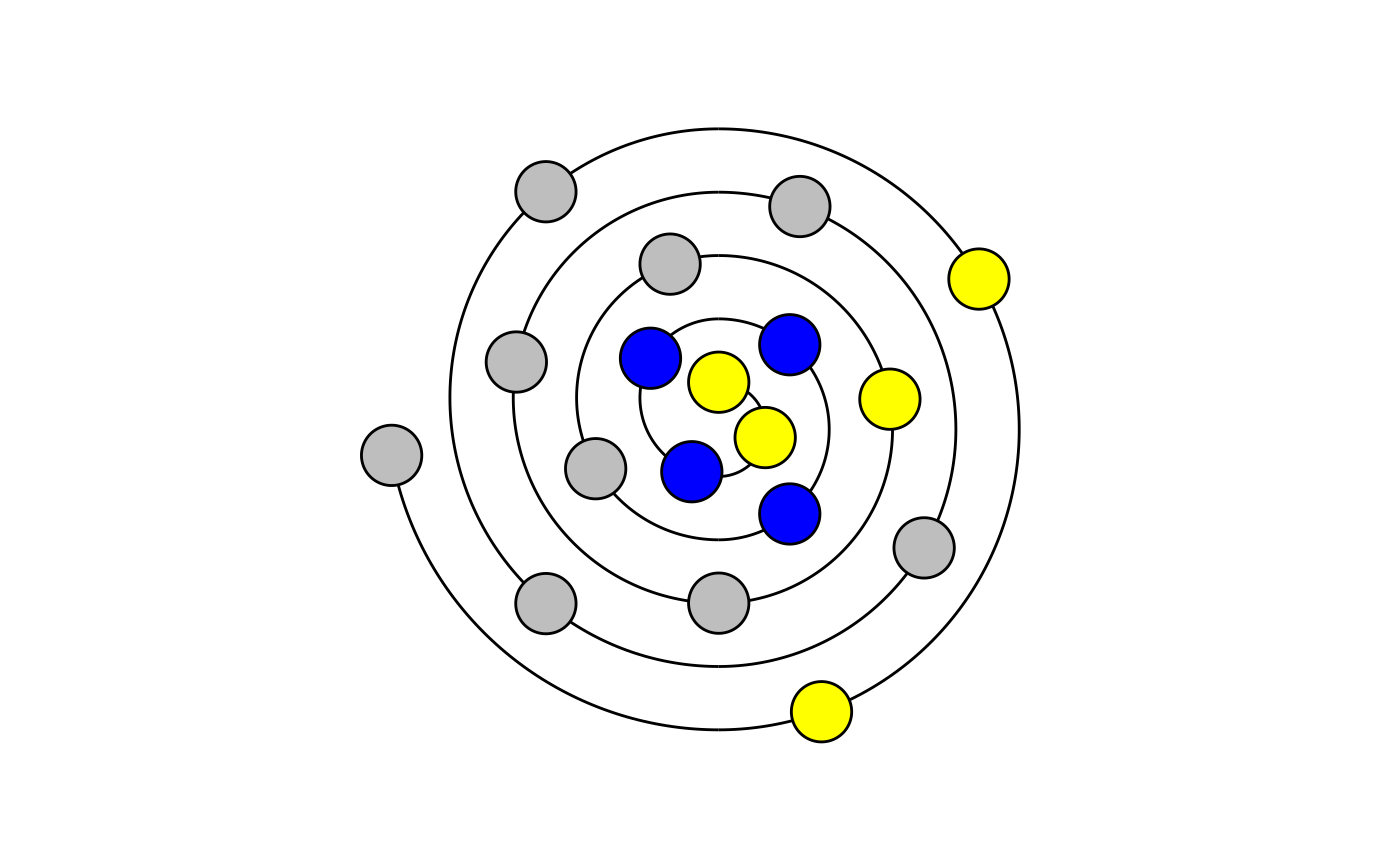
Create Wenxiang diagrams to visualize alpha helical sequences.
Source:R/draw_wenxiang.R
draw_wenxiang.RdThis function visualizes alpha-helical peptides as Wenxiang diagrams. Sequences between 2 and 18 (inclusive) characters can be visualized. The residue closest to the center represents the amino acid at the N-terminus (first in `sequence`).
draw_wenxiang(sequence, col = c("grey", "yellow", "blue", "red"), labels = FALSE, label.col = "black", fixed = TRUE, legend = FALSE)
Arguments
| sequence | character vector containing amino acid sequence from N-terminus to C-terminus |
|---|---|
| col | colors for each amino acid type in the following order: nonpolar residues, polar residues, basic residues, acidic residues |
| labels | logical value; if TRUE, one-letter residue codes are overlaid on the residue circles |
| label.col | character value for color of labels added if `labels = TRUE` |
| fixed | if TRUE (default), ensures that residues will be circles (not ellipses) even if graphics device is rectangular |
| legend | if TRUE, adds legend to plot |
Examples
draw_wenxiang("GIGAVLKVLTTGLPALIS")draw_wenxiang("QQRKRKIWSILAPLGTTL")